Friday, July 15, 2022
Perturb-Seq in Our Database
Tuesday, July 5, 2022
Crispr-i Vs Conventional Knockdown
It is now possible to read out the transcriptional alterations induced by alteration of every gene in the genome in a single experiment. Actually, this has been possible since the advent of microarrays, but recent advances in crispr and single-cell sequencing technologies means the feat can be accomplished without gargantuan expenditures. Crispr inhibition ("crispr knockdown" or "crispri") clogs up the promoter of any gene of interest. Bar codes allow the experimenter to know which gene has been clogged. And single-cell sequencing gives you the transcriptome of a clogged cell. After intensive computation, you get a simple matrix of genes knocked down and genes altered.
Above is a very small matrix. The yellow column shows genes that are knocked down. The blue row shows genes examined for alterations. The matrix needn't be square...in the study we'll be talking about, there were more genes examined than genes knocked down. So, you can see that knocking down pd-l1 strongly upregulates tubb in this very hypothetical example. Here, the gene of interest is found in the yellow column. But the gene of interest can also be found in the blue row. So, for example, you can ask, "What are the genes whose knockdown upregulates ifn?" There, pd-l1 knockdown does the best job of upregulating ifn. This is the approach taken in crispr and rnai experiments that ask questions like, "What genes most strongly control the expression of cancer migratory factor XYZ?"
The study of interest is here. K562 cells were used. Roughly 8,000 genes were knocked down, and 10,000 genes were examined for transcriptional alterations. Typically, 100 cells were examined for a particular knockdown, meaning that more than a million single cells were sequenced. We'll be adding the data to our database, but we need to make a few tweaks to our system to accomodate the data. However, we have some preliminary results of combining this "perturb-seq" dataset with our own data. Below, we discuss these results. They're weird.
First, a bit more background. Crispri is often referred to as a "knockdown." The gene of interest hasn't been mutated or deleted. A protein and RNA guide simply clog up a promoter. Thus, as with standard rnai, knockdown can be of varying efficiency. In both forms of knockdown, off-target effects are also possible. However, crispri differs (profoundly, we believe) from standard rnai in that the knockdown occurs right at the DNA/RNA interface, at the level of transcription, while rnai functions after full-length transcripts have already been generated. Rnai results in by-products of cleaved RNA and/or protein-bound transcripts with inhibited translation.
Despite the differences in these two knockdown approaches, it seems reasonable to expect that both sorts of knockdowns should have similar transcriptomic effects. In both cases, simple RT-PCR can tell you that the gene of interest has been stymied by, say, 90%. In both cases, there isn't much transcript floating around for translation into protein. Nevertheless, experimental comparisons between the approaches are necessary. Unfortunately, we don't see extensive examples of such comparisons. Probably the best study is found here. Surprisingly, even in a controlled setting using a single cell line (Hela), the transcriptomic readouts of crispri and rnai did not match up particularly well. We note another possible comparison here. The comparison is less than ideal, however, since the experimenters abandoned crispri in favor of rnai because of less-than-desirable knockdown efficiency.
To more closely examine the discrepancies between crispri and more conventional means of altering gene expression (knockdown/out, overexpression, drug targeting, mutation), we first sought out the most commonly altered genes in our database. TP53 ranks first, with a total of 45 studies targeting it. Unfortunately, TP53 was not strongly downregulated in the k562 perturb-seq data on which we operated; this particular crispri knockdown was not particularly effective.
The next most commonly targeted gene in the database is TGF-b, with a total of 36 studies. The gene is indeed the single-most strongly downregulated gene upon TGF-b perturb-seq knockdown. However, we have a new problem in comparing conventional studies against crispri; all but 3 of the above studies involve treatment. That is, cells are bathed in TGF-b. It would not be fair to expect perturb-seq results to parallel these treatment results. Nevertheless, we note the following:
*The single conventional TGF-b knockdown study actually mimicked TGF treatment fairly well, albeit in opposite directions (i.e. genes upregulated in the knockdown are downregulated upon treatment, and vice-versa).
*Perturb-seq results did not match up with any significance with ANY of the above 36 studies.
Given the time we spent tinkering with TGF-b data, we constructed lists of genes that are canonically up/down-regulated on TGF-b treatment: database IDs 145169203 and 145170203 (to be available upon our next database update).
EZH2 is targeted 32 times in our database. This time, it is strongly downregulated on crispri treatment. We also have a variety of knockdown/out, overexpression, mutation, drug-targeting results relating to EZH2. Do any of these studies at all match up with the perturb-seq results?
No.
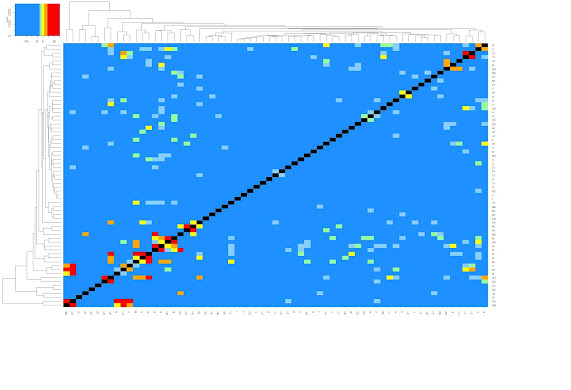
To be fair, there's not a lot of study/study overlap here. Presumably, that's because the effects of EZH2 alteration depend strongly on cell type. Below is a heat map showing EZH2 study/study P-values. Bear in mind that the underlying gene lists are divided into up- and down-regulated portions...even in a best case scenario, half of the lists aren't expected to overlap. Nevertheless, green/yellow/orange/red color makes it clear that most studies overlap with at least one other study with decent significance. The up/down lists for perturb-seq, N2 and O2, however, lie smack in the least colorful columns/rows of the heatmap. In fact, the single best EZH2-related match for perturb-seq is this: genes that are upregulated under perturb-seq crispri tend to be downregulated under standard EZH2 KO in an MBA-MB-231 line (log(P)=-2.8, see GSE48979).
Stuff that might be true
I'll add to the below list as thoughts pop into my brain.... *"Celebrity" genes are over-rated. Last I looked there are someth...
-
"T-Cell Exhaustion" is associated with an inability of the immune system to fight off cancer and other diseases. We grabbed 7 mark...
-
Here, we discuss the use of WIMG tools to search for drugs or treatments or gene perturbations that may reverse various disease signatures....
-
Most of the gene lists that compose our database are sorted according to some criteria. We may apply a significance cut-off to the data, and...